ELECTROCHEMICAL BIOSCAFFOLDS FOR THE DETERMINATION OF HIGHLY RELEVANT BIOMARKERS IN DIAGNOSIS AND PROGNOSIS OF CANCER: MIRNAS
|
Description |
The offered technology allows reliable, selective and sensitive determination of early diagnosis related cancer biomarkers (microRNAs, miRNAs or miRs) in total RNA extracts (RNAt) from complex clinical samples, in a rapid and simple manner without requirement for additional amplification steps, by employment of amperometric biosensors based on magnetic micro-carriers functionalized with suitable bioreceptors and disposable screen printed electrodes as electrochemical transducers.
The recorded amperometric signal, based on enzymatic and electrochemical reactions involved in the HRP/HQ/H2O2 system, that occur on the surface of the electrochemical transducer at a fixed potential, is directly proportional to the target miRNA concentration contained in the sample to be analyzed.
|
How does it work |
The fundamentals of the developed amperometric platform are based on the selective hybridization between the target miRNA and its fully complementary probe, which is modified with biotin in one extreme of the sequence, and the subsequent capture of the above hybrids, formed previously in solution, by high selective bioreceptors that have been effectively immobilized onto the surface of commercial magnetic micro-carriers. Finally, the captured hybrids are enzymatically labeled with Streptavidin-HRP polymer through biotin-streptavidin interaction.
The developed amperometric devices have been successfully applied to the determination of several mature miRNAs which are highly relevant for diagnosis and prognosis of breast cancer, such as miRNA-21 and miRNA-205, in RNAt extracts from cell lines, fresh and paraffin embedded breast tissues and breast cytologies, just by supplementing the appropriate quantity of the RNAt extracted from the sample with the specific biotinylated complementary probe.
Due to absence of matrix effect in all tested samples, mature miRNA concentration in a given sample can be simply determined by interpolating the obtained amperometric signal in a calibration plot performed with synthetic target miRNA standards.
Although the optimized time for determination of target miRNA ranges from 75 to 120 minutes, it is possible to reduce drastically the assay time to only 15 minutes without a significant loss of sensitivity.
Because of the high sequence homology existing between different miRNAs contained simultaneously in a given sample, methodologies for analyzing these types of biomarkers must possess great selectivity. The developed methodologies have demonstrated high selectivity against non-complementary miRNAs sequences and an acceptable selectivity against 1-mismatched (1-m) miRNAs sequences, which ensures reliable and accurate results. Moreover, the high versatility of the proposed methodology makes it easily transferable to the determination of any miRNA and to the multiplexed analysis of a panel of miRNAs, allowing the characterization, at a tissue and cell level, of miRNAs expression profiles that are inherent to any type and stage of the disease. In this sense, application of these devices for characterization of miRNAs expression profiles could permit to perform the molecular classification of the disease and subsequently provide the most appropriate clinical treatment in each case.
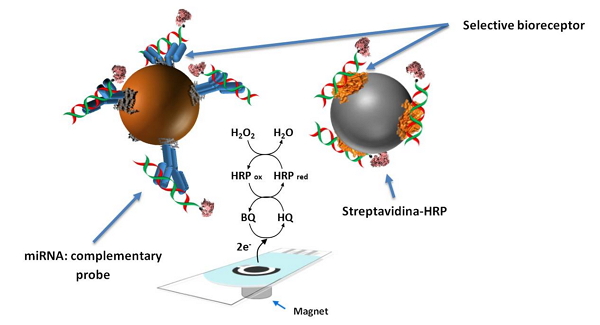
Schematic display of the developed electrochemical bioscaffolds for miRNAs determination.
|
Advantages |
- Some of the main advantages of the proposed methodology are:
- Possible applicability for the detection of any miRNA and other types of RNAs in RNAt extracted from any type of biological sample
- Simple implementation as “point-of-care” diagnostics devices
- Simple and cost-effective instrumentation
- Enough sensitivity for direct detection of miRNAs with no need of additional tedious steps, such as reverse transcription, amplification and/or purification
- Multiplexing capabilities
- Easy automated technology
- Lower assay times and costs in comparison with conventional methodologies for miRNAs analysis (qRT-PCR)
- Provide reliable quantitative results
- Similar efficiency obtained in the analysis of miRNAs in fresh and paraffin embedded tissues (FFPE)
- Possibility to perform the analysis at room temperature
|
Where has it been developed |
These electrochemical bioplatforms have been performed in the group of Electroanalysis and (Bio) Electrochemical sensors at the Faculty of Chemistry at the Complutense University of Madrid, headed by Prof. José Manuel Pingarrón in collaboration with the group of Molecular Pharmacology from CIB-CSIC (Dr. José María Sánchez-Puelles) and CANNAN RESEARCH & INVESTMENT S.L company (D. Enrique Sáinz-Martínez).
|
And also |
The offered methodology has been also successfully applied to the individual and simultaneous detection of two relevant breast cancer miRNAs: miRNA-21 and miRNA-205. These two target miRNAs show opposite roles and expression levels in breast cancer; over-expression or oncogene role for miRNA-21 and down-expression or tumor suppressor role for miRNA-205.
The developed methodologies allow obtaining absolute concentrations of target miRNAs in a simpler, rapid and cost-effective manner in comparison to qRT-PCR, the gold standard method for miRNA detection. In addition, these methodologies avoid the requirements of previous and tedious reverse transcription steps from RNA to DNA, as well as amplification and/or the use of internal standards.
Both the great operation and the high degree of robustness demonstrated by the proposed strategies allow their implementation as powerful analytical tools able to provide additional information to the most frequently clinical test for breast cancer diagnosis, based on semiquantitative assays of protein biomarkers expression levels (immunohistochemistry).
|
Contact |
|
© Office for the Transfer of Research Results – UCM |
|
PDF Downloads |
|
Classification |
|
Responsible Researchers |
José Manuel Pingarrón Carrazón: pingarro@quim.ucm.es
Susana Campuzano Ruiz: susanacr@quim.ucm.es
Department: Analytical Chemistry
Faculty: Chemistry Sciences